Tiny Nanopore Sensor: Imagine being able to unlock the secrets of your DNA with a device small enough to fit in your pocket. Thanks to tiny nanopore sensors, this is no longer just a dream for scientists in high-tech labs—it’s rapidly becoming a reality for doctors, researchers, and even students around the globe. This revolutionary technology is changing the way we approach genetics, making DNA sequencing faster, more affordable, and accessible to everyone.
Tiny Nanopore Sensor
DNA sequencing is the process of reading the unique code that makes up all living things. For decades, this process was slow, expensive, and limited to specialized laboratories. Now, with the advent of nanopore sensors, we are witnessing a dramatic shift. These sensors are not only tiny and portable, but they also provide real-time results and require minimal sample preparation. This means that DNA sequencing can be done almost anywhere—from a hospital bedside to a rainforest, or even in space.
The implications are enormous. Faster and more affordable DNA sequencing can lead to quicker disease diagnosis, more precise treatments, better outbreak tracking, and new discoveries in fields like agriculture, conservation, and forensics.
Tiny Nanopore Sensor
| Feature/Fact | Details & Examples |
|---|---|
| Technology | Nanopore sequencing uses tiny holes (nanopores) to read DNA/RNA directly, in real time. |
| Speed | Can deliver results in real time; whole genome sequencing possible in hours, not days. |
| Cost | Devices are portable and much cheaper than traditional sequencers; consumables also less expensive. |
| Accuracy | Recent advances have improved accuracy, including detection of modified bases. |
| Applications | Used for clinical diagnostics, outbreak surveillance, environmental monitoring, and more. |
| Official Website | Oxford Nanopore Technologies |
Tiny nanopore sensors are revolutionizing DNA sequencing by making it faster, cheaper, and more accessible than ever before. Their real-time, portable, and user-friendly design is opening new possibilities in medicine, research, and education. With ongoing improvements in accuracy and speed, nanopore sequencing is set to become a cornerstone of modern science and healthcare.
What Is Nanopore Sequencing?

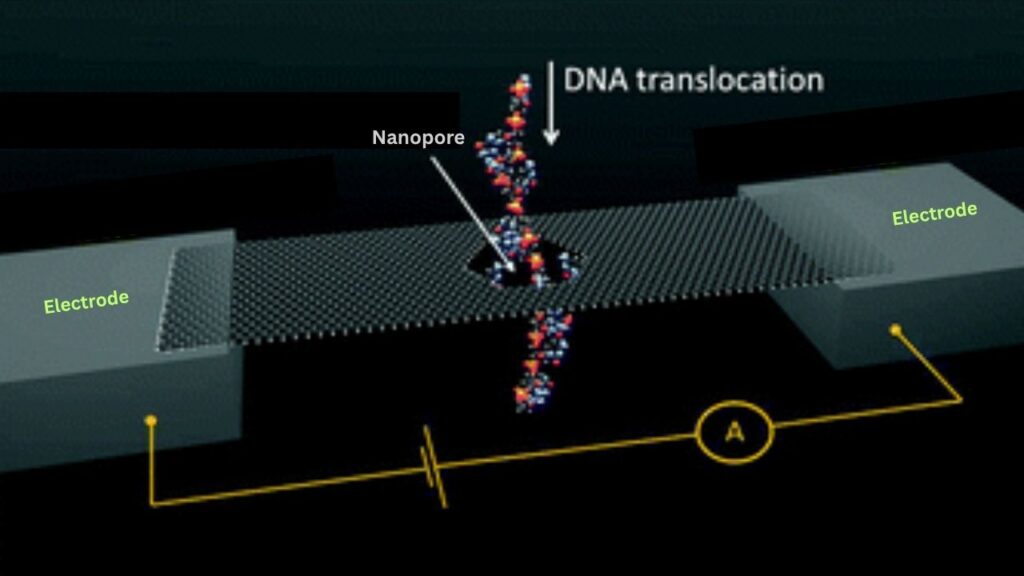
Nanopore sequencing is a method for reading the sequence of DNA or RNA molecules using a nanoscale hole—called a nanopore—embedded in a thin membrane. As a strand of DNA or RNA passes through the nanopore, it disrupts an electrical current in a unique way. Special software translates these disruptions into the sequence of genetic letters (A, T, C, G for DNA; A, U, C, G for RNA).
How Is It Different from Traditional Sequencing?
Traditional sequencing methods, such as Sanger or next-generation sequencing (NGS), require complex sample preparation, expensive equipment, and can take days to deliver results. Nanopore sequencing, however, can read long stretches of DNA in real time, with minimal preparation, and on devices that are portable and affordable.
Key Advantages
- Direct, label-free detection: Reads native DNA or RNA, including chemical modifications, without the need for amplification or labeling.
- Portability: Devices like the MinION are small and lightweight, making them ideal for fieldwork and point-of-care diagnostics.
- Scalability: The technology can be used for small, targeted studies or scaled up for large population genomics projects.
- Real-time analysis: Results are available as soon as sequencing begins, allowing for rapid decision-making.
How Does Nanopore Sequencing Work? (A Step-by-Step Guide)
1. Sample Preparation
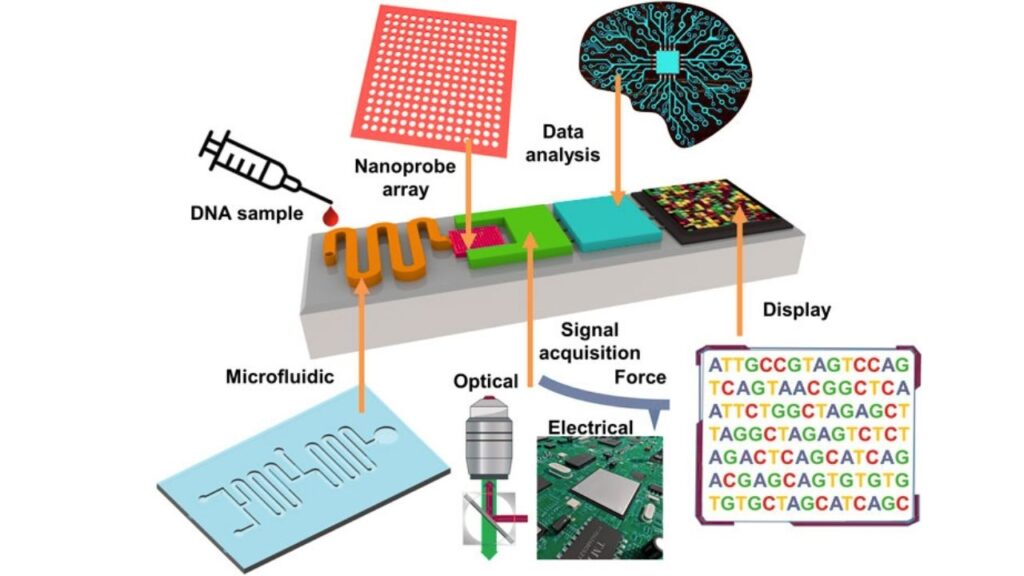
The first step is to extract DNA or RNA from the sample. This could be blood, saliva, plant tissue, or even environmental samples like soil or water. The extracted genetic material is then prepared using special kits that attach adapters and a motor protein to the ends of the DNA or RNA strands. The motor protein controls the speed at which the strand moves through the nanopore.
2. Loading the Flow Cell
The prepared sample is loaded into a flow cell, which contains thousands of nanopores embedded in a membrane. Each nanopore acts as a tiny sensor.
3. Sequencing
An electric current is applied across the membrane. As each DNA or RNA strand is drawn through a nanopore by the motor protein, it causes characteristic disruptions in the current. These disruptions are detected by sensors and sent to a computer.
4. Data Analysis
The computer software interprets the changes in current to identify the sequence of bases. This process happens in real time, so users can monitor the progress and quality of the sequencing as it happens.
5. Interpretation and Application
Once the sequence data is generated, it can be analyzed for a variety of purposes—identifying pathogens, detecting genetic mutations, studying biodiversity, or developing personalized medicine strategies.
Real-World Applications of Nanopore Sequencing
Disease Outbreaks and Public Health
Nanopore sequencing has been a game-changer in tracking infectious diseases. During the COVID-19 pandemic, portable nanopore devices were used to sequence the virus’s genome directly at outbreak sites. This enabled rapid identification of new variants, informed public health decisions, and helped contain the spread of the virus.
Clinical Diagnostics
Doctors and clinical laboratories are using nanopore sequencing to quickly identify bacteria, viruses, and fungi in patient samples. This is especially valuable for detecting antibiotic resistance genes, allowing for more targeted and effective treatments. In some cases, diagnosis that once took days can now be completed in a matter of hours.
Environmental and Agricultural Research
Researchers use nanopore sequencing to study microbes in soil, water, and air. This helps monitor pollution, track the spread of invasive species, and ensure food safety. In agriculture, nanopore sequencing can identify plant pathogens and support the development of disease-resistant crops.
Conservation and Wildlife Biology
Conservationists use portable nanopore sequencers to analyze DNA from endangered species, even in remote locations. This information helps track genetic diversity, monitor populations, and develop strategies for species recovery.
Personalized Medicine
As sequencing costs drop, it becomes feasible to sequence an individual’s genome as part of routine healthcare. Doctors can use this information to predict disease risk, choose the most effective medications, and design personalized treatment plans.
Nanopore Sequencing vs. Traditional Sequencing
| Feature | Nanopore Sequencing | Traditional Sequencing (e.g., Illumina) |
|---|---|---|
| Read Length | Ultra-long (up to millions of bases) | Short (typically 150–300 bases) |
| Speed | Real-time, hours | Batch processing, days |
| Portability | Handheld devices available | Large, stationary lab equipment |
| Sample Prep | Minimal, no amplification needed | Requires amplification and labeling |
| Cost | Lower device and running costs | Higher initial and operational costs |
| Direct Detection | Yes (native DNA/RNA, modifications) | No (often reads amplified, labeled DNA) |
The Science Behind Nanopore Sensors
Nanopore sensors are built using advanced materials, including biological proteins and synthetic membranes. The nanopore itself is just a few nanometers wide—about 100,000 times smaller than the width of a human hair. The sensitivity of the sensor allows it to distinguish between the four DNA bases as they pass through the pore.
Recent advances include the use of solid-state nanopores made from materials like graphene and molybdenum disulfide. These are more durable and easier to manufacture at scale, paving the way for even more affordable and reliable devices.
Practical Guide: Getting Started with Nanopore Sequencing
1. Define Your Project
Decide what you want to sequence. Are you interested in a specific gene, an entire genome, or a mix of organisms in a sample? Nanopore technology is flexible and can handle a wide range of projects.
2. Choose the Right Device
- MinION: Ideal for small labs, fieldwork, and educational use.
- GridION and PromethION: Suitable for high-throughput projects and large-scale studies.
3. Prepare Your Sample
Follow the manufacturer’s protocols for DNA/RNA extraction and library preparation. Many commercial kits are available to simplify this process.
4. Run the Sequencer
Load your prepared sample into the flow cell, start the device, and monitor the sequencing run using the provided software. Data is available in real time.
5. Analyze and Interpret Data
Use bioinformatics tools to process the raw data. Many user-friendly software options exist, from basic sequence analysis to advanced genomics workflows.
6. Apply Your Results
Interpret the findings in the context of your research or clinical question. For example, identify disease-causing mutations, track the spread of a pathogen, or assess biodiversity in an ecosystem.
Ensuring Data Quality and Accuracy
While nanopore sequencing is highly innovative, maintaining data accuracy is critical. Improvements in nanopore chemistry, motor proteins, and signal processing algorithms have significantly increased accuracy rates. Users are encouraged to:
- Use high-quality DNA/RNA samples.
- Follow best practices for library preparation.
- Utilize updated software for basecalling and error correction.
- Validate findings with complementary methods when necessary, especially for clinical decisions.
New Drug Delivery Breakthrough Uses Molecular Switches to Control When Medicine Works in the Body
New Discovery in Atomic Physics Could Unlock Powerful Quantum Behaviors in Ultra-Cold Matter
Could Trilayer Graphene’s Unusually High Kinetic Inductance Revolutionize Nanoelectronics?
FAQs About Tiny Nanopore Sensor
Q1: Is nanopore sequencing accurate enough for clinical use?
A: Yes, accuracy has improved greatly in recent years. With proper protocols and software, nanopore sequencing can provide clinically actionable results. For some applications, results may be confirmed with other methods.
Q2: How much does it cost to use a nanopore sequencer?
A: Entry-level devices like the MinION are available for a few thousand dollars, with consumables priced for routine use. This is significantly less expensive than traditional sequencers, which can cost hundreds of thousands of dollars.
Q3: Can nanopore sequencing be used outside the lab?
A: Absolutely. Nanopore devices are designed for portability and can be used in clinics, field stations, and remote locations. They have even been used on the International Space Station.
Q4: What are the main limitations of nanopore sequencing?
A: While accuracy and throughput have improved, some applications may still benefit from complementary sequencing technologies. Long-read sequencing can also be sensitive to sample quality.
Q5: Who uses nanopore sequencing today?
A: Researchers, clinicians, public health officials, conservationists, and educators worldwide use nanopore sequencing for a wide range of applications.
The Future of DNA Sequencing: Accessible to All
The tiny nanopore sensor is more than just a technical innovation—it’s a tool for democratizing access to genetic information. As technology advances, we can expect even faster, more accurate, and more affordable sequencing. This will enable personalized healthcare, rapid outbreak response, improved crop breeding, and new discoveries in biology.
Imagine a future where anyone, anywhere, can read and understand the code of life—unlocking new possibilities for science, medicine, and society.